Articles on Statistics and Machine Learning for Healthcare
- linear and generalized linear models

Linear Regression Summary(lm): Interpretting in R
Introduction to linear regression summary printouts.
In this post we describe how to interpret the summary of a linear regression model in R given by summary(lm). We discuss interpretation of the residual quantiles and summary statistics, the standard errors and t statistics , along with the p-values of the latter, the residual standard error, and the F-test. Let’s first load the Boston housing dataset and fit a naive model. We won’t worry about assumptions, which are described in other posts.
Residual Summary Statistics
We can investigate this further with a boxplot of the residuals.
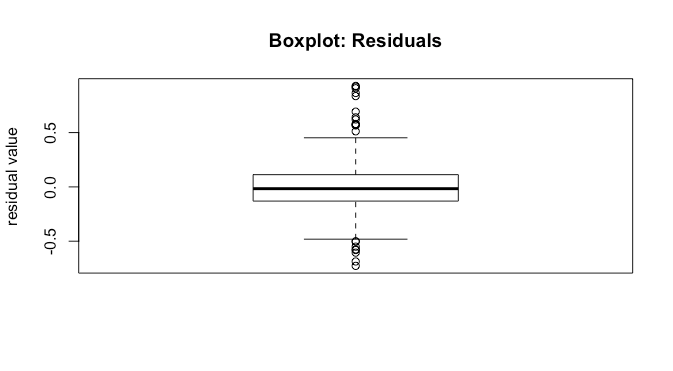
Coefficients
The second thing printed by the linear regression summary call is information about the coefficients. This includes their estimates, standard errors, t statistics, and p-values.
Standard Error
- your model assumptions hold
Here we can see that the entire confidence interval for number of rooms has a large effect size relative to the other covariates.
The t-statistic is
Pr(>|t|)
Assessing Fit and Overall Significance
Residual Standard Error
Multiple and Adjusted
F-Statistic and F-test
In addition to looking at whether individual features have a significant effect, we may also wonder whether at least one feature has a significant effect. That is, we would like to test the null hypothesis
Related Posts
Linear regression: comparing models between two groups with linearhypothesis september 21, 2019 why you should center your features in linear regression august 31, 2019 leave a reply cancel reply.
Your email address will not be published. Required fields are marked *
This site uses Akismet to reduce spam. Learn how your comment data is processed .
Learn by Marketing
Data mining + marketing in plain english.
- Data Science Reading List
Explaining the lm() Summary in R
- Residual Standard Error : Essentially standard deviation of residuals / errors of your regression model.
- Multiple R-Squared : Percent of the variance of Y intact after subtracting the error of the model.
- Adjusted R-Squared : Same as multiple R-Squared but takes into account the number of samples and variables you’re using.
- F-Statistic : Global test to check if your model has at least one significant variable. Takes into account number of variables and observations used.
R’s lm() function is fast, easy, and succinct. However, when you’re getting started, that brevity can be a bit of a curse. I’m going to explain some of the key components to the summary() function in R for linear regression models. In addition, I’ll also show you how to calculate these figures for yourself so you have a better intuition of what they mean.
Getting Started: Build a Model
Before we can examine a model summary, we need to build a model. To follow along with this example, create these three variables.
Just for fun, I’m using data from Anscombe’s quartet (Q1) and then creating a second variable with a defined pattern and some random error.
Now, we’ll create a linear regression model using R’s lm() function and we’ll get the summary output using the summary() function.
This is the output you should receive.
Meaning Behind Each Section of Summary()
I’m not going to focus on the Call, Residuals, or Coefficients section. If you’re doing regression analysis, you should understand residuals and the coefficient section. Here’s a brief description of each as a refresher.
- Call : This is an R feature that shows what function and parameters were used to create the model.
- Residuals : Difference between what the model predicted and the actual value of y. You can calculate the Residuals section like so: summary(y-model$fitted.values)
- Std. Error is Residual Standard Error (see below) divided by the square root of the sum of the square of that particular x variable.
- t value: Estimate divided by Std. Error
- Pr(>|t|): Look up your t value in a T distribution table with the given degrees of freedom.
With those sections out of the way, we’ll focus on the bottom of the summary output.
Residual Standard Error
In R, the lm summary produces the standard deviation of the error with a slight twist. Standard deviation is the square root of variance. Standard Error is very similar. The only difference is that instead of dividing by n-1, you subtract n minus 1 + # of variables involved.
Multiple R-Squared
Also called the coefficient of determination, this is an oft-cited measurement of how well your model fits to the data. While there are many issues with using it alone (see Anscombe’s quartet ) , it’s a quick and pre-computed check for your model.
R-Squared subtracts the residual error from the variance in Y. The bigger the error, the worse the remaining variance will appear.
If you notice, numerator doesn’t have to be positive. If the model is so bad, you can actually end up with a negative R-Squared.
Adjusted R-Squared
Multiple R-Squared works great for simple linear (one variable) regression. However, in most cases, the model has multiple variables. The more variables you add, the more variance you’re going to explain. So you have to control for the extra variables.
Adjusted R-Squared normalizes Multiple R-Squared by taking into account how many samples you have and how many variables you’re using.
Notice how k is in the denominator. If you have 100 observations (n) and 5 variables, you’ll be dividing by 100-5-1 = 94. If you have 20 variables instead, you’re dividing by 100-20-1 = 79. As the denominator gets smaller, the results get larger: 99 /94 = 1.05; 79/94 = 1.25.
A larger normalizing value is going to make the Adjusted R-Squared worse since we’re subtracting its product from one.
F-Statistic
Finally, the F-Statistic. Including the t-tests, this is the second “test” that the summary function produces for lm models. The F-Statistic is a “global” test that checks if at least one of your coefficients are nonzero.
The reason for this test is based on the fact that if you run multiple hypothesis tests (namely, on your coefficients), you’re likely to include a variable that isn’t actually significant. See this for an example (and an explanation ).
You can now replicate the summary statistics produced by R’s summary function on linear regression (lm) models!
If you’re interested in more R tutorials on linear regression and beyond, take a look at the Linear Regression page.
Quick Guide: Interpreting Simple Linear Model Output in R
Linear regression models are a key part of the family of supervised learning models.

Linear regression models are a key part of the family of supervised learning models. In particular, linear regression models are a useful tool for predicting a quantitative response. For more details, check an article I’ve written on Simple Linear Regression - An example using R . In general, statistical softwares have different ways to show a model output. This quick guide will help the analyst who is starting with linear regression in R to understand what the model output looks like. In the example below, we’ll use the cars dataset found in the datasets package in R (for more details on the package you can call: library(help = "datasets") .
The cars dataset gives Speed and Stopping Distances of Cars. This dataset is a data frame with 50 rows and 2 variables. The rows refer to cars and the variables refer to speed (the numeric Speed in mph) and dist (the numeric stopping distance in ft.). As the summary output above shows, the cars dataset’s speed variable varies from cars with speed of 4 mph to 25 mph (the data source mentions these are based on cars from the ’20s! - to find out more about the dataset, you can type ?cars ). When it comes to distance to stop, there are cars that can stop in 2 feet and cars that need 120 feet to come to a stop.
Below is a scatterplot of the variables:
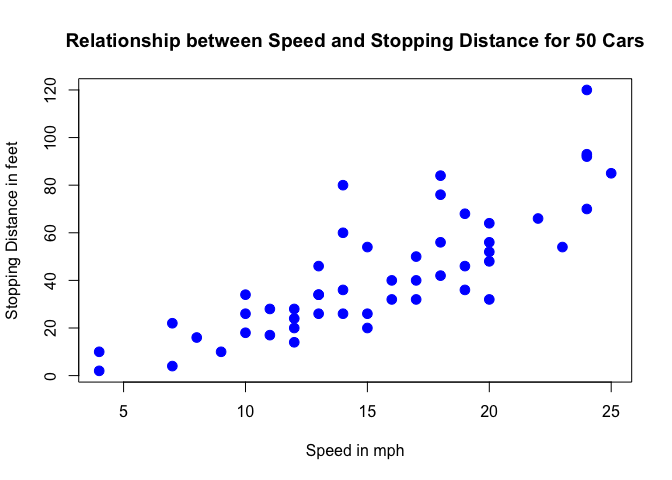
From the plot above, we can visualise that there is a somewhat strong relationship between a cars’ speed and the distance required for it to stop (i.e.: the faster the car goes the longer the distance it takes to come to a stop).
In this exercise, we will:
- Run a simple linear regression model in R and distil and interpret the key components of the R linear model output. Note that for this example we are not too concerned about actually fitting the best model but we are more interested in interpreting the model output - which would then allow us to potentially define next steps in the model building process.
Let’s get started by running one example:
The model above is achieved by using the lm() function in R and the output is called using the summary() function on the model.
Below we define and briefly explain each component of the model output:
Formula Call
As you can see, the first item shown in the output is the formula R used to fit the data. Note the simplicity in the syntax: the formula just needs the predictor (speed) and the target/response variable (dist), together with the data being used (cars).
The next item in the model output talks about the residuals. Residuals are essentially the difference between the actual observed response values (distance to stop dist in our case) and the response values that the model predicted. The Residuals section of the model output breaks it down into 5 summary points. When assessing how well the model fit the data, you should look for a symmetrical distribution across these points on the mean value zero (0). In our example, we can see that the distribution of the residuals do not appear to be strongly symmetrical. That means that the model predicts certain points that fall far away from the actual observed points. We could take this further consider plotting the residuals to see whether this normally distributed, etc. but will skip this for this example.
Coefficients
The next section in the model output talks about the coefficients of the model. Theoretically, in simple linear regression, the coefficients are two unknown constants that represent the intercept and slope terms in the linear model. If we wanted to predict the Distance required for a car to stop given its speed, we would get a training set and produce estimates of the coefficients to then use it in the model formula. Ultimately, the analyst wants to find an intercept and a slope such that the resulting fitted line is as close as possible to the 50 data points in our data set.
Coefficient - Estimate
The coefficient Estimate contains two rows; the first one is the intercept. The intercept, in our example, is essentially the expected value of the distance required for a car to stop when we consider the average speed of all cars in the dataset. In other words, it takes an average car in our dataset 42.98 feet to come to a stop. The second row in the Coefficients is the slope, or in our example, the effect speed has in distance required for a car to stop. The slope term in our model is saying that for every 1 mph increase in the speed of a car, the required distance to stop goes up by 3.9324088 feet.
Coefficient - Standard Error
The coefficient Standard Error measures the average amount that the coefficient estimates vary from the actual average value of our response variable. We’d ideally want a lower number relative to its coefficients. In our example, we’ve previously determined that for every 1 mph increase in the speed of a car, the required distance to stop goes up by 3.9324088 feet. The Standard Error can be used to compute an estimate of the expected difference in case we ran the model again and again. In other words, we can say that the required distance for a car to stop can vary by 0.4155128 feet. The Standard Errors can also be used to compute confidence intervals and to statistically test the hypothesis of the existence of a relationship between speed and distance required to stop.
Coefficient - t value
The coefficient t-value is a measure of how many standard deviations our coefficient estimate is far away from 0. We want it to be far away from zero as this would indicate we could reject the null hypothesis - that is, we could declare a relationship between speed and distance exist. In our example, the t-statistic values are relatively far away from zero and are large relative to the standard error, which could indicate a relationship exists. In general, t-values are also used to compute p-values.
Coefficient - Pr(>t)
The Pr(>t) acronym found in the model output relates to the probability of observing any value equal or larger than t . A small p-value indicates that it is unlikely we will observe a relationship between the predictor (speed) and response (dist) variables due to chance. Typically, a p-value of 5% or less is a good cut-off point. In our model example, the p-values are very close to zero. Note the ‘signif. Codes’ associated to each estimate. Three stars (or asterisks) represent a highly significant p-value. Consequently, a small p-value for the intercept and the slope indicates that we can reject the null hypothesis which allows us to conclude that there is a relationship between speed and distance.
Residual Standard Error
Residual Standard Error is measure of the quality of a linear regression fit. Theoretically, every linear model is assumed to contain an error term E . Due to the presence of this error term, we are not capable of perfectly predicting our response variable (dist) from the predictor (speed) one. The Residual Standard Error is the average amount that the response (dist) will deviate from the true regression line. In our example, the actual distance required to stop can deviate from the true regression line by approximately 15.3795867 feet, on average. In other words, given that the mean distance for all cars to stop is 42.98 and that the Residual Standard Error is 15.3795867 , we can say that the percentage error is (any prediction would still be off by) 35.78% . It’s also worth noting that the Residual Standard Error was calculated with 48 degrees of freedom. Simplistically, degrees of freedom are the number of data points that went into the estimation of the parameters used after taking into account these parameters (restriction). In our case, we had 50 data points and two parameters (intercept and slope).
Multiple R-squared, Adjusted R-squared
The R-squared ($R^2$) statistic provides a measure of how well the model is fitting the actual data. It takes the form of a proportion of variance. $R^2$ is a measure of the linear relationship between our predictor variable (speed) and our response / target variable (dist). It always lies between 0 and 1 (i.e.: a number near 0 represents a regression that does not explain the variance in the response variable well and a number close to 1 does explain the observed variance in the response variable). In our example, the $R^2$ we get is 0.6510794. Or roughly 65% of the variance found in the response variable (dist) can be explained by the predictor variable (speed). Step back and think: If you were able to choose any metric to predict distance required for a car to stop, would speed be one and would it be an important one that could help explain how distance would vary based on speed? I guess it’s easy to see that the answer would almost certainly be a yes. That why we get a relatively strong $R^2$. Nevertheless, it’s hard to define what level of $R^2$ is appropriate to claim the model fits well. Essentially, it will vary with the application and the domain studied.
A side note: In multiple regression settings, the $R^2$ will always increase as more variables are included in the model. That’s why the adjusted $R^2$ is the preferred measure as it adjusts for the number of variables considered.
F-Statistic
F-statistic is a good indicator of whether there is a relationship between our predictor and the response variables. The further the F-statistic is from 1 the better it is. However, how much larger the F-statistic needs to be depends on both the number of data points and the number of predictors. Generally, when the number of data points is large, an F-statistic that is only a little bit larger than 1 is already sufficient to reject the null hypothesis (H0 : There is no relationship between speed and distance). The reverse is true as if the number of data points is small, a large F-statistic is required to be able to ascertain that there may be a relationship between predictor and response variables. In our example the F-statistic is 89.5671065 which is relatively larger than 1 given the size of our data.
Note that the model we ran above was just an example to illustrate how a linear model output looks like in R and how we can start to interpret its components. Obviously the model is not optimised. One way we could start to improve is by transforming our response variable (try running a new model with the response variable log-transformed mod2 = lm(formula = log(dist) ~ speed.c, data = cars) or a quadratic term and observe the differences encountered). We could also consider bringing in new variables, new transformation of variables and then subsequent variable selection, and comparing between different models. Finally, with a model that is fitting nicely, we could start to run predictive analytics to try to estimate distance required for a random car to stop given its speed.
You May Also Like...
- Tips for Creating Better-Looking PowerBI Dashboards
- Bad vs Good Data Viz: A Simple Bar Chart Example
- Reimagining Data Science: The Rise of LLM-Powered Agents
- Data-Driven Solutions to Help You Elevate Your Marketing Efforts Today
- Mastering Data Visualisation and Storytelling: A Guide for Leaders
- © Felipe Rego
- Inspired by Massively Theme
- Design by HTML5 UP
- Jekyll Integration by Somiibo
lm: Fitting Linear Models
Description.
lm is used to fit linear models. It can be used to carry out regression, single stratum analysis of variance and analysis of covariance (although aov may provide a more convenient interface for these).
an object of class " formula " (or one that can be coerced to that class): a symbolic description of the model to be fitted. The details of model specification are given under ‘Details’.
an optional data frame, list or environment (or object coercible by as.data.frame to a data frame) containing the variables in the model. If not found in data , the variables are taken from environment(formula) , typically the environment from which lm is called.
an optional vector specifying a subset of observations to be used in the fitting process.
an optional vector of weights to be used in the fitting process. Should be NULL or a numeric vector. If non-NULL, weighted least squares is used with weights weights (that is, minimizing sum(w*e^2) ); otherwise ordinary least squares is used. See also ‘Details’,
a function which indicates what should happen when the data contain NA s. The default is set by the na.action setting of options , and is na.fail if that is unset. The ‘factory-fresh’ default is na.omit . Another possible value is NULL , no action. Value na.exclude can be useful.
the method to be used; for fitting, currently only method = "qr" is supported; method = "model.frame" returns the model frame (the same as with model = TRUE , see below).
logicals. If TRUE the corresponding components of the fit (the model frame, the model matrix, the response, the QR decomposition) are returned.
logical. If FALSE (the default in S but not in R ) a singular fit is an error.
an optional list. See the contrasts.arg of model.matrix.default .
this can be used to specify an a priori known component to be included in the linear predictor during fitting. This should be NULL or a numeric vector or matrix of extents matching those of the response. One or more offset terms can be included in the formula instead or as well, and if more than one are specified their sum is used. See model.offset .
additional arguments to be passed to the low level regression fitting functions (see below).
lm returns an object of class "lm" or for multiple responses of class c("mlm", "lm") .
The functions summary and anova are used to obtain and print a summary and analysis of variance table of the results. The generic accessor functions coefficients , effects , fitted.values and residuals extract various useful features of the value returned by lm .
An object of class "lm" is a list containing at least the following components:
a named vector of coefficients
the residuals, that is response minus fitted values.
the fitted mean values.
the numeric rank of the fitted linear model.
(only for weighted fits) the specified weights.
the residual degrees of freedom.
the matched call.
the terms object used.
(only where relevant) the contrasts used.
(only where relevant) a record of the levels of the factors used in fitting.
the offset used (missing if none were used).
if requested, the response used.
if requested, the model matrix used.
if requested (the default), the model frame used.
(where relevant) information returned by model.frame on the special handling of NA s.
In addition, non-null fits will have components assign, effects and (unless not requested) qr relating to the linear fit, for use by extractor functions such as summary and effects .
Using time series
Considerable care is needed when using lm with time series.
Unless na.action = NULL , the time series attributes are stripped from the variables before the regression is done. (This is necessary as omitting NA s would invalidate the time series attributes, and if NA s are omitted in the middle of the series the result would no longer be a regular time series.)
Even if the time series attributes are retained, they are not used to line up series, so that the time shift of a lagged or differenced regressor would be ignored. It is good practice to prepare a data argument by ts.intersect (…, dframe = TRUE) , then apply a suitable na.action to that data frame and call lm with na.action = NULL so that residuals and fitted values are time series.
Models for lm are specified symbolically. A typical model has the form response ~ terms where response is the (numeric) response vector and terms is a series of terms which specifies a linear predictor for response . A terms specification of the form first + second indicates all the terms in first together with all the terms in second with duplicates removed. A specification of the form first:second indicates the set of terms obtained by taking the interactions of all terms in first with all terms in second . The specification first*second indicates the cross of first and second . This is the same as first + second + first:second .
If the formula includes an offset , this is evaluated and subtracted from the response.
If response is a matrix a linear model is fitted separately by least-squares to each column of the matrix.
See model.matrix for some further details. The terms in the formula will be re-ordered so that main effects come first, followed by the interactions, all second-order, all third-order and so on: to avoid this pass a terms object as the formula (see aov and demo(glm.vr) for an example).
A formula has an implied intercept term. To remove this use either y ~ x - 1 or y ~ 0 + x . See formula for more details of allowed formulae.
Non- NULL weights can be used to indicate that different observations have different variances (with the values in weights being inversely proportional to the variances); or equivalently, when the elements of weights are positive integers \(w_i\), that each response \(y_i\) is the mean of \(w_i\) unit-weight observations (including the case that there are \(w_i\) observations equal to \(y_i\) and the data have been summarized). However, in the latter case, notice that within-group variation is not used. Therefore, the sigma estimate and residual degrees of freedom may be suboptimal; in the case of replication weights, even wrong. Hence, standard errors and analysis of variance tables should be treated with care.
lm calls the lower level functions lm.fit , etc, see below, for the actual numerical computations. For programming only, you may consider doing likewise.
All of weights , subset and offset are evaluated in the same way as variables in formula , that is first in data and then in the environment of formula .
Chambers, J. M. (1992) Linear models. Chapter 4 of Statistical Models in S eds J. M. Chambers and T. J. Hastie, Wadsworth & Brooks/Cole.
Wilkinson, G. N. and Rogers, C. E. (1973). Symbolic descriptions of factorial models for analysis of variance. Applied Statistics , 22 , 392--399. 10.2307/2346786.
summary.lm for summaries and anova.lm for the ANOVA table; aov for a different interface.
The generic functions coef , effects , residuals , fitted , vcov .
predict.lm (via predict ) for prediction, including confidence and prediction intervals; confint for confidence intervals of parameters .
lm.influence for regression diagnostics, and glm for generalized linear models.
The underlying low level functions, lm.fit for plain, and lm.wfit for weighted regression fitting.
More lm() examples are available e.g., in anscombe , attitude , freeny , LifeCycleSavings , longley , stackloss , swiss .
biglm in package biglm for an alternative way to fit linear models to large datasets (especially those with many cases).
Run the code above in your browser using DataCamp Workspace

R news and tutorials contributed by hundreds of R bloggers
Beautiful tables for linear model summaries #rstats.
Posted on March 6, 2015 by Daniel in R bloggers | 0 Comments
[social4i size="small" align="align-left"] --> [This article was first published on Strenge Jacke! » R , and kindly contributed to R-bloggers ]. (You can report issue about the content on this page here ) Want to share your content on R-bloggers? click here if you have a blog, or here if you don't.
Beautiful HTML tables of linear models
In this blog post I’d like to show some (old and) new features of the sjt.lm function from my sjPlot-package . These functions are currently only implemented in the development snapshot on GitHub . A package update is planned to be submitted soon to CRAN.
There are two new major features I added to this function: Comparing models with different predictors (e.g. stepwise regression) and automatic grouping of categorical predictors. There are examples below that demonstrate these features.
The sjt.lm function prints results and summaries of linear models as HTML-table. These tables can be viewed in the RStudio Viewer pane, web browser or easily exported to office applications. See also my former posts on the table printing functions of my package here and here .
Please note: The following tables may look a bit cluttered – this is because I just pasted the HTML-code created by knitr into this blog post, so style sheets may interfere. The original online-manual for this function can be found here .
All following tables can be reproduced with the sjPlot package and the sample data set from this package.
Linear model summaries as HTML table
The sjt.lm function prints summaries of linear models (fitted with the lm function) as nicely formatted html-tables.
Before starting, sample data is loaded and sample models are fitted:
The simplest way of producing the table output is by passing the fitted models as parameter. By default, estimates ( B ), confidence intervals ( CI ) and p-values ( p ) are reported. The models are named Model 1 and Model 2 .
Custom labels
You can specify the ‘model’ label via labelDependentVariables parameter:
More custom labels
Here is an example how to change the other labels. Note that showHeaderStrings makes the two labels on top and top left corner appear in the table.
Changing summary style and content
You can change the table style with specific parameters, e.g. to include CI into the same table cell as the estimates, print asterisks instead of numeric p-values etc.
Custom variable labels
In the above example, the original variable labels are long and not much pretty. You can change variable labels either with set_var_labels (see this page for more detaila), which will affect all future plots and tables, or pass own labels via labelPredictors .
Compare models with different predictors
In some cases, for instance stepwise regressions, you have different predictors on the same response. The proper grouping of predictors, resp. rows, is done automatically.
First, let’s fit some example models.
Note that printing tables with fitted models, which have different predictors do not automatically detect variable labels (maybe this will be implemented in a future package version).
Automatic grouping of categorical predictors
In case you have categorical variables with more than two factor levels, the sjt.lm function automatically groups the category levels to give a better overview of predictors in the table.
By default, automatic grouping is activated. To disable this feature, use group.pred = FALSE as parameter.
To demonstrate this feature, we first convert two predictors to factors (what they actually are, indeed). To do this, we use the to_fac function, which converts numerical variables into factors, however, does not remove the variable and value label attributes.
Now we can print the table.
Removing estimates from the output
With remove.estmates , specific estimates can be removed from the table output. This may make sense in case you have stepwise regression models and only want to compare the varying predictors but not the controls. remove.estmates either accepts the row indices of the rows of the table output that should be removed, or the coefficient’s names.
When using numeric indices, the estimates’ index number relates to the same order as coef(fit) . Note that currently the intercept cannot be removed from the model output!
Example 1: Complete table output
Here you have the complete table output. This helps you identify the row index numbers. Especially when you have multiple models with different predictors, the estimate’s position in the last model may differ from this estimate’s position in the table output.

Example 2: Remove first coefficient (after intercept)
Example 3: remove age and sex, example 4: remove many esimates, example 5: custom predictor labels.
In most cases you need to define your own labels when removing estimates, especially when you have grouped categorical predictors, because automatic label detection in quite tricky in such situations. If you provide own labels, please note that grouped predictors’ headings (the variable name of the grouped, categorical variable) are still automatically set by the sjt.lm function (variable labels are used, so use set_var_labels for those categorical predictors). All data rows in the table, i.e. for each coefficient appearing in the model, you need to specify a label string.
In the next example, we have seven table rows with data (excluding intercept): mid and hi education (categories of the variable Education), Hours of Care, slight, moderate and severe dependency (categories of the variable Dependency) and Service Usage. These ‘rows’ need to be labelled.
To leave a comment for the author, please follow the link and comment on their blog: Strenge Jacke! » R . R-bloggers.com offers daily e-mail updates about R news and tutorials about learning R and many other topics. Click here if you're looking to post or find an R/data-science job . Want to share your content on R-bloggers? click here if you have a blog, or here if you don't.
Copyright © 2022 | MH Corporate basic by MH Themes
Never miss an update! Subscribe to R-bloggers to receive e-mails with the latest R posts. (You will not see this message again.)
Have a language expert improve your writing
Run a free plagiarism check in 10 minutes, generate accurate citations for free.
- Knowledge Base
- Linear Regression in R | A Step-by-Step Guide & Examples
Linear Regression in R | A Step-by-Step Guide & Examples
Published on February 25, 2020 by Rebecca Bevans . Revised on June 22, 2023.
Linear regression is a regression model that uses a straight line to describe the relationship between variables . It finds the line of best fit through your data by searching for the value of the regression coefficient(s) that minimizes the total error of the model.
There are two main types of linear regression:
- Simple linear regression uses only one independent variable
- Multiple linear regression uses two or more independent variables
In this step-by-step guide, we will walk you through linear regression in R using two sample datasets.
Download the sample datasets to try it yourself.
Simple regression dataset Multiple regression dataset
Table of contents
Getting started in r, step 1: load the data into r, step 2: make sure your data meet the assumptions, step 3: perform the linear regression analysis, step 4: check for homoscedasticity, step 5: visualize the results with a graph, step 6: report your results, other interesting articles.
Start by downloading R and RStudio . Then open RStudio and click on File > New File > R Script .
As we go through each step , you can copy and paste the code from the text boxes directly into your script. To run the code, highlight the lines you want to run and click on the Run button on the top right of the text editor (or press ctrl + enter on the keyboard).
To install the packages you need for the analysis, run this code (you only need to do this once):
Next, load the packages into your R environment by running this code (you need to do this every time you restart R):
Prevent plagiarism. Run a free check.
Follow these four steps for each dataset:
- In RStudio, go to File > Import dataset > From Text (base) .
- Choose the data file you have downloaded ( income.data or heart.data ), and an Import Dataset window pops up.
- In the Data Frame window, you should see an X (index) column and columns listing the data for each of the variables ( income and happiness or biking , smoking , and heart.disease ).
- Click on the Import button and the file should appear in your Environment tab on the upper right side of the RStudio screen.
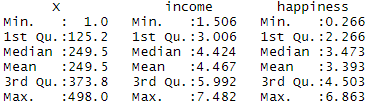
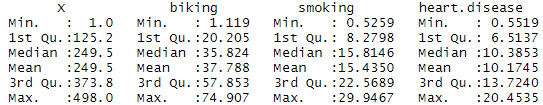
After you’ve loaded the data, check that it has been read in correctly using summary() .
Simple regression
Because both our variables are quantitative , when we run this function we see a table in our console with a numeric summary of the data. This tells us the minimum, median , mean , and maximum values of the independent variable (income) and dependent variable (happiness):
Multiple regression
Again, because the variables are quantitative, running the code produces a numeric summary of the data for the independent variables (smoking and biking) and the dependent variable (heart disease):
We can use R to check that our data meet the four main assumptions for linear regression .
- Independence of observations (aka no autocorrelation)
Because we only have one independent variable and one dependent variable, we don’t need to test for any hidden relationships among variables.
If you know that you have autocorrelation within variables (i.e. multiple observations of the same test subject), then do not proceed with a simple linear regression! Use a structured model, like a linear mixed-effects model, instead.
To check whether the dependent variable follows a normal distribution , use the hist() function.
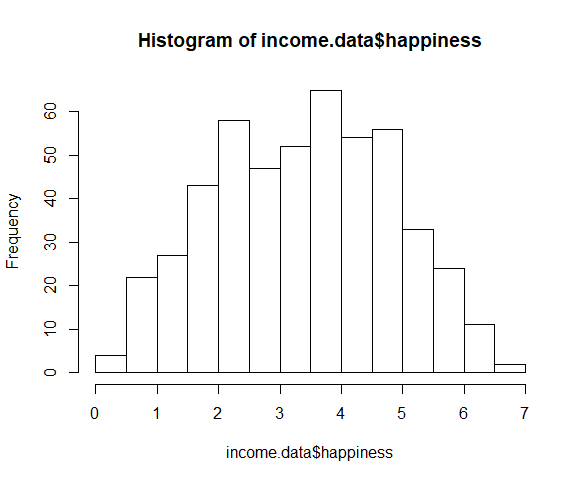
The observations are roughly bell-shaped (more observations in the middle of the distribution, fewer on the tails), so we can proceed with the linear regression.
The relationship between the independent and dependent variable must be linear. We can test this visually with a scatter plot to see if the distribution of data points could be described with a straight line.
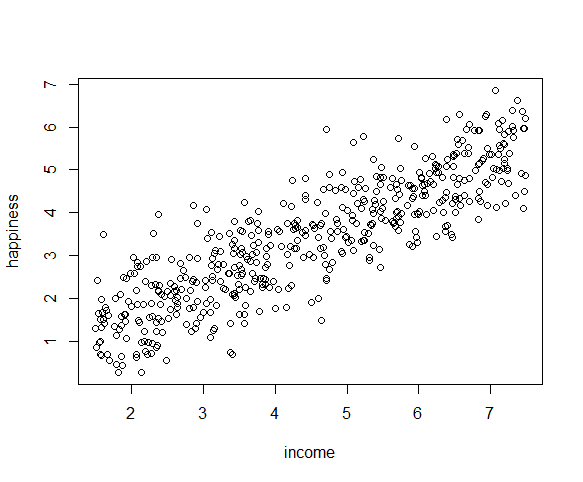
The relationship looks roughly linear, so we can proceed with the linear model.
- Homoscedasticity (aka homogeneity of variance )
This means that the prediction error doesn’t change significantly over the range of prediction of the model. We can test this assumption later, after fitting the linear model.
Use the cor() function to test the relationship between your independent variables and make sure they aren’t too highly correlated.
When we run this code, the output is 0.015. The correlation between biking and smoking is small (0.015 is only a 1.5% correlation), so we can include both parameters in our model.
Use the hist() function to test whether your dependent variable follows a normal distribution .
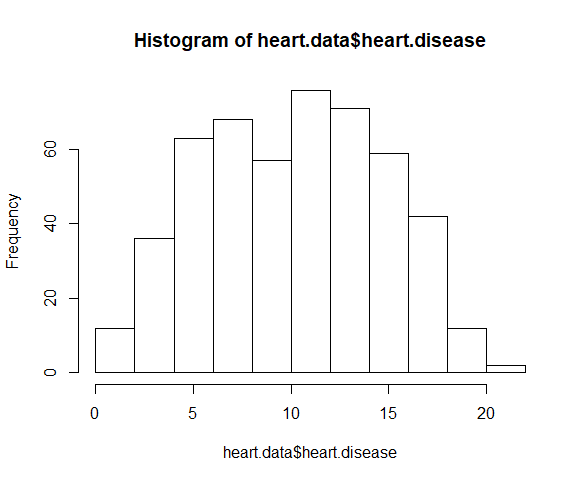
The distribution of observations is roughly bell-shaped, so we can proceed with the linear regression.
We can check this using two scatterplots: one for biking and heart disease, and one for smoking and heart disease.
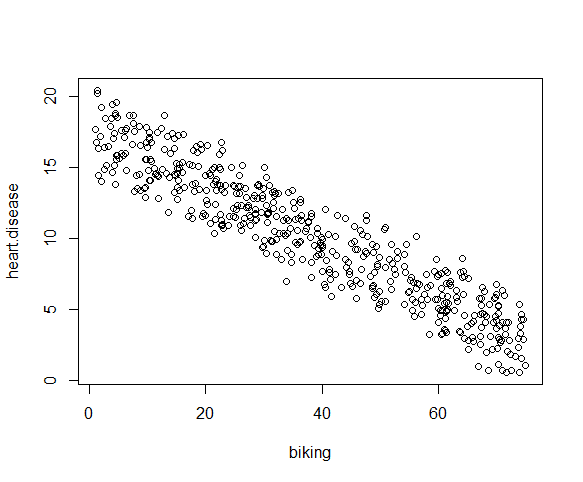
Although the relationship between smoking and heart disease is a bit less clear, it still appears linear. We can proceed with linear regression.
- Homoscedasticity
We will check this after we make the model.
Now that you’ve determined your data meet the assumptions, you can perform a linear regression analysis to evaluate the relationship between the independent and dependent variables.
Simple regression: income and happiness
Let’s see if there’s a linear relationship between income and happiness in our survey of 500 people with incomes ranging from $15k to $75k, where happiness is measured on a scale of 1 to 10.
To perform a simple linear regression analysis and check the results, you need to run two lines of code. The first line of code makes the linear model, and the second line prints out the summary of the model:
The output looks like this:
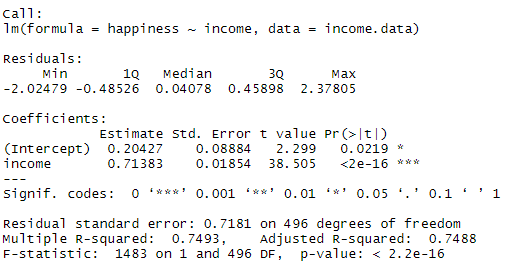
This output table first presents the model equation, then summarizes the model residuals (see step 4).
The Coefficients section shows:
- The estimates ( Estimate ) for the model parameters – the value of the y-intercept (in this case 0.204) and the estimated effect of income on happiness (0.713).
- The standard error of the estimated values ( Std. Error ).
- The test statistic ( t value , in this case the t statistic ).
- The p value ( Pr(>| t | ) ), aka the probability of finding the given t statistic if the null hypothesis of no relationship were true.
The final three lines are model diagnostics – the most important thing to note is the p value (here it is 2.2e-16, or almost zero), which will indicate whether the model fits the data well.
From these results, we can say that there is a significant positive relationship between income and happiness ( p value < 0.001), with a 0.713-unit (+/- 0.01) increase in happiness for every unit increase in income.
Multiple regression: biking, smoking, and heart disease
Let’s see if there’s a linear relationship between biking to work, smoking, and heart disease in our imaginary survey of 500 towns. The rates of biking to work range between 1 and 75%, rates of smoking between 0.5 and 30%, and rates of heart disease between 0.5% and 20.5%.
To test the relationship, we first fit a linear model with heart disease as the dependent variable and biking and smoking as the independent variables. Run these two lines of code:
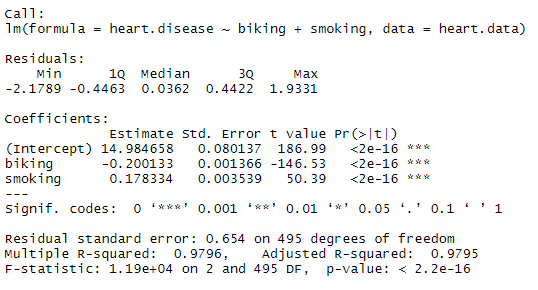
The estimated effect of biking on heart disease is -0.2, while the estimated effect of smoking is 0.178.
This means that for every 1% increase in biking to work, there is a correlated 0.2% decrease in the incidence of heart disease. Meanwhile, for every 1% increase in smoking, there is a 0.178% increase in the rate of heart disease.
The standard errors for these regression coefficients are very small, and the t statistics are very large (-147 and 50.4, respectively). The p values reflect these small errors and large t statistics. For both parameters, there is almost zero probability that this effect is due to chance.
Remember that these data are made up for this example, so in real life these relationships would not be nearly so clear!
Receive feedback on language, structure, and formatting
Professional editors proofread and edit your paper by focusing on:
- Academic style
- Vague sentences
- Style consistency
See an example
Before proceeding with data visualization, we should make sure that our models fit the homoscedasticity assumption of the linear model.
We can run plot(income.happiness.lm) to check whether the observed data meets our model assumptions:
Note that the par(mfrow()) command will divide the Plots window into the number of rows and columns specified in the brackets. So par(mfrow=c(2,2)) divides it up into two rows and two columns. To go back to plotting one graph in the entire window, set the parameters again and replace the (2,2) with (1,1).
These are the residual plots produced by the code:
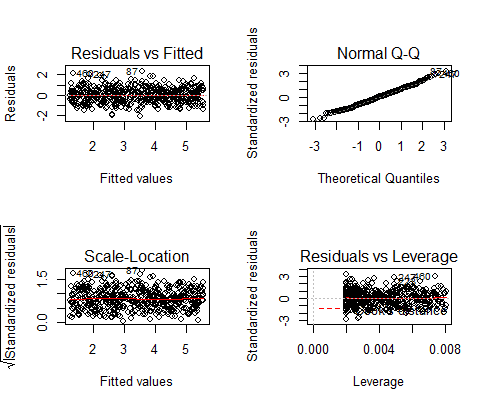
Residuals are the unexplained variance . They are not exactly the same as model error, but they are calculated from it, so seeing a bias in the residuals would also indicate a bias in the error.
The most important thing to look for is that the red lines representing the mean of the residuals are all basically horizontal and centered around zero. This means there are no outliers or biases in the data that would make a linear regression invalid.
In the Normal Q-Qplot in the top right, we can see that the real residuals from our model form an almost perfectly one-to-one line with the theoretical residuals from a perfect model.
Based on these residuals, we can say that our model meets the assumption of homoscedasticity.
Again, we should check that our model is actually a good fit for the data, and that we don’t have large variation in the model error, by running this code:
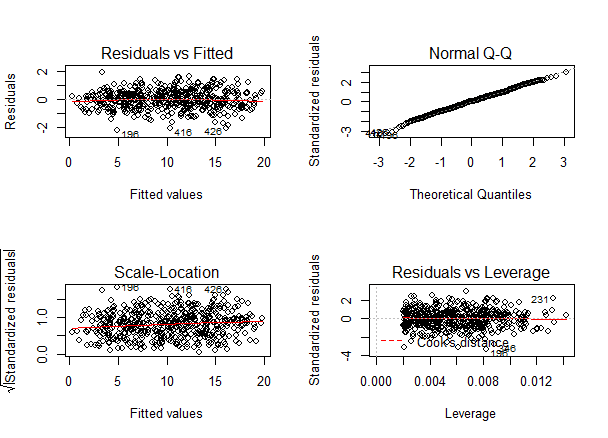
As with our simple regression, the residuals show no bias, so we can say our model fits the assumption of homoscedasticity.
Next, we can plot the data and the regression line from our linear regression model so that the results can be shared.
Follow 4 steps to visualize the results of your simple linear regression.
- Plot the data points on a graph
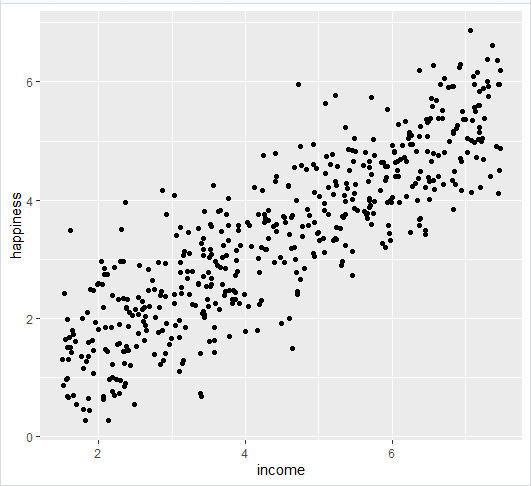
- Add the linear regression line to the plotted data
Add the regression line using geom_smooth() and typing in lm as your method for creating the line. This will add the line of the linear regression as well as the standard error of the estimate (in this case +/- 0.01) as a light grey stripe surrounding the line:
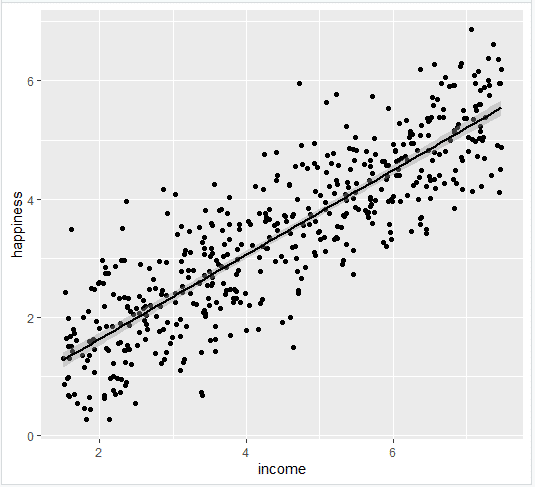
- Add the equation for the regression line.
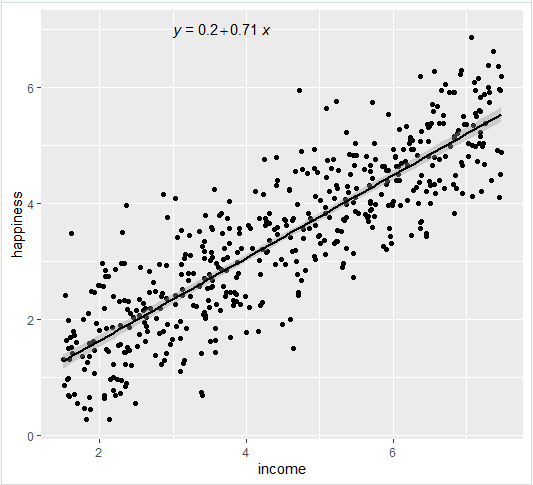
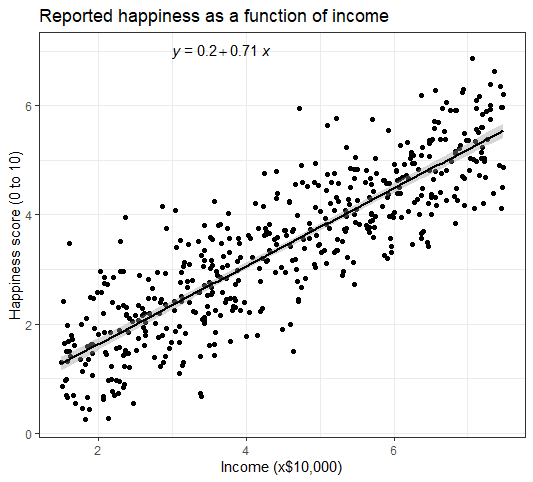
- Make the graph ready for publication
We can add some style parameters using theme_bw() and making custom labels using labs() .
This produces the finished graph that you can include in your papers:
The visualization step for multiple regression is more difficult than for simple regression, because we now have two predictors. One option is to plot a plane, but these are difficult to read and not often published.
We will try a different method: plotting the relationship between biking and heart disease at different levels of smoking. In this example, smoking will be treated as a factor with three levels, just for the purposes of displaying the relationships in our data.
There are 7 steps to follow.
- Create a new dataframe with the information needed to plot the model
Use the function expand.grid() to create a dataframe with the parameters you supply. Within this function we will:
- Create a sequence from the lowest to the highest value of your observed biking data;
- Choose the minimum, mean, and maximum values of smoking, in order to make 3 levels of smoking over which to predict rates of heart disease.
This will not create anything new in your console, but you should see a new data frame appear in the Environment tab. Click on it to view it.
- Predict the values of heart disease based on your linear model
Next we will save our ‘predicted y’ values as a new column in the dataset we just created.
- Round the smoking numbers to two decimals
This will make the legend easier to read later on.
- Change the ‘smoking’ variable into a factor
This allows us to plot the interaction between biking and heart disease at each of the three levels of smoking we chose.
- Plot the original data
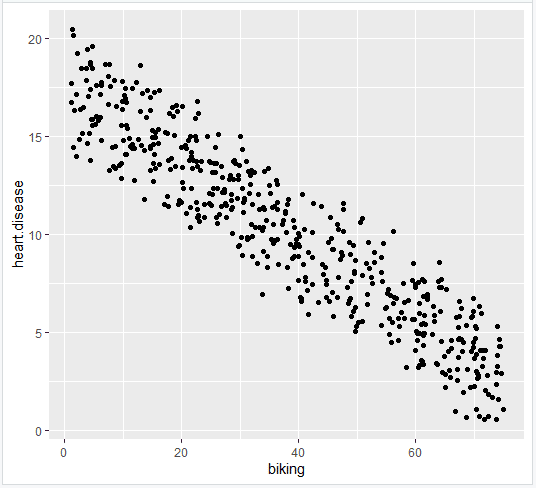
- Add the regression lines
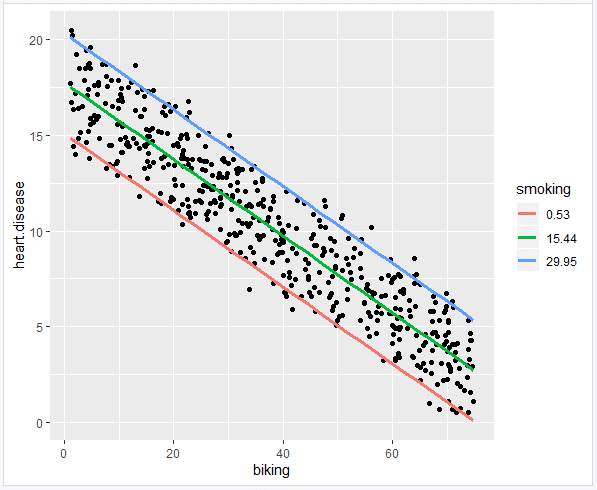
Because this graph has two regression coefficients, the stat_regline_equation() function won’t work here. But if we want to add our regression model to the graph, we can do so like this:
This is the finished graph that you can include in your papers!
In addition to the graph, include a brief statement explaining the results of the regression model.
If you want to know more about statistics , methodology , or research bias , make sure to check out some of our other articles with explanations and examples.
- Chi square test of independence
- Statistical power
- Descriptive statistics
- Degrees of freedom
- Pearson correlation
- Null hypothesis
Methodology
- Double-blind study
- Case-control study
- Research ethics
- Data collection
- Hypothesis testing
- Structured interviews
Research bias
- Hawthorne effect
- Unconscious bias
- Recall bias
- Halo effect
- Self-serving bias
- Information bias
Cite this Scribbr article
If you want to cite this source, you can copy and paste the citation or click the “Cite this Scribbr article” button to automatically add the citation to our free Citation Generator.
Bevans, R. (2023, June 22). Linear Regression in R | A Step-by-Step Guide & Examples. Scribbr. Retrieved April 2, 2024, from https://www.scribbr.com/statistics/linear-regression-in-r/
Is this article helpful?
Rebecca Bevans
Other students also liked, simple linear regression | an easy introduction & examples, multiple linear regression | a quick guide (examples), choosing the right statistical test | types & examples, what is your plagiarism score.
summary.lm: Summarizing Linear Model Fits
Summarizing linear model fits, description.
summary method for class "lm" .
print.summary.lm tries to be smart about formatting the coefficients, standard errors, etc. and additionally gives ‘significance stars’ if signif.stars is TRUE .
Aliased coefficients are omitted in the returned object but restored by the print method.
Correlations are printed to two decimal places (or symbolically): to see the actual correlations print summary(object)$correlation directly.
The function summary.lm computes and returns a list of summary statistics of the fitted linear model given in object , using the components (list elements) "call" and "terms" from its argument, plus
The model fitting function lm , summary .
Function coef will extract the matrix of coefficients with standard errors, t-statistics and p-values.
R Package Documentation
Browse r packages, we want your feedback.

Add the following code to your website.
REMOVE THIS Copy to clipboard
For more information on customizing the embed code, read Embedding Snippets .
An Introduction to R
6.3 simple linear modelling.
Linear models are one of the most widely used models in statistics and data science. They are often thought of as simple models but they’re very flexible and able to model a wide variety of experimental and survey designs. Many of the statistical approaches you may have used previously (such as linear regression, t -test, ANOVA, ANCOVA etc) can be expressed as a linear model so the good news is that you’re probably already familiar with linear models (albeit indirectly). They also form the foundation of more complicated modelling approaches and are relatively easy to extended to incorporate additional complexity. During this section we’ll learn how to fit some simple linear models using R and cover some of the more common applications. We won’t go into any detail of the underlying linear modelling theory but rather focus on the practicalities of model fitting and R code.
The main function for fitting linear models in R is the lm() function (short for linear model!). The lm() function has many arguments but the most important is the first argument which specifies the model you want to fit using a model formula which typically takes the general form:
response variable ~ explanatory variable(s)
This model formula is simply read as
‘variation in the response variable modelled as a function (~) of the explanatory variable(s)’.
The response variable is also commonly known as the ‘dependent variable’ and the explanatory variables are sometimes referred to as ‘independent variables’ (or less frequently as ‘predictor variables’). There is also an additional term in our model formula which represents the variation in our response variable not explained by our explanatory variables but you don’t need to specify this when using the lm() function.
As mentioned above, many of the statistical ‘tests’ you might have previously used can be expressed as a linear model. For example, if we wanted to perform a bivariate linear regression between a response variable ( y ) and a single continuous explanatory variable ( x ) our model formula would simply be
On the other hand, if we wanted to use an ANOVA to test whether the group means of a response variable ( y ) were different between a three level factor ( x ) our model formula would look like
OK, hang on, they both look identical, what gives? In addition to the model formula, the type of linear model you fit is also determined by the type of data in your explanatory variable(s) (i.e. what class of data). If your explanatory variable is continuous then you will fit a bivariate linear regression. If your explanatory variable is a factor (i.e. categorical data) you will fit an ANOVA type model.
You can also increase the complexity of your linear model by including additional explanatory variables in your model formula. For example, if we wanted to fit a two-way ANOVA both of our explanatory variables x and z would need to be factors and separated by a + symbol
If we wanted to perform a factorial ANOVA to identify an interaction between both explanatory variables we would separate our explanatory variables with a : symbol whilst also including our main effects in our model formula
or by using the equivalent shortcut notation
It’s important that you get comfortable with using model formula (and we’ve only given the briefest of explanations above) when using the lm() function (and other functions) as it’s remarkably easy to specifiy a model which is either nonsense or isn’t the model you really wanted to fit. A summary table of various linear model formula and equivalent R code given below.
OK, time for an example. The data file smoking.txt summarises the results of a study investigating the possible relationship between mortality rate and smoking across 25 occupational groups in the UK. The variable occupational.group specifies the different occupational groups studied, the risk.group variable indicates the relative risk to lung disease for the various occupational groups and smoking is an index of the average number of cigarettes smoked each day (relative to the number smoked across all occupations). The variable mortality is an index of the death rate from lung cancer in each group (relative to the death rate across all occupational groups). In this data set, the response variable is mortality and the potential explanatory variables are smoking which is numeric and risk.group which is a three level factor. The first thing to do is import our data file using the read.table() function as usual and assign the data to an object called smoke . You can find a link to download these data here .
Next, let’s investigate the relationship between the mortality and smoking variables by plotting a scatter plot. We can use either the ggplot2 package or base R graphics to do this. We’ll use ggplot2 this time and our old friend the ggplot() function.
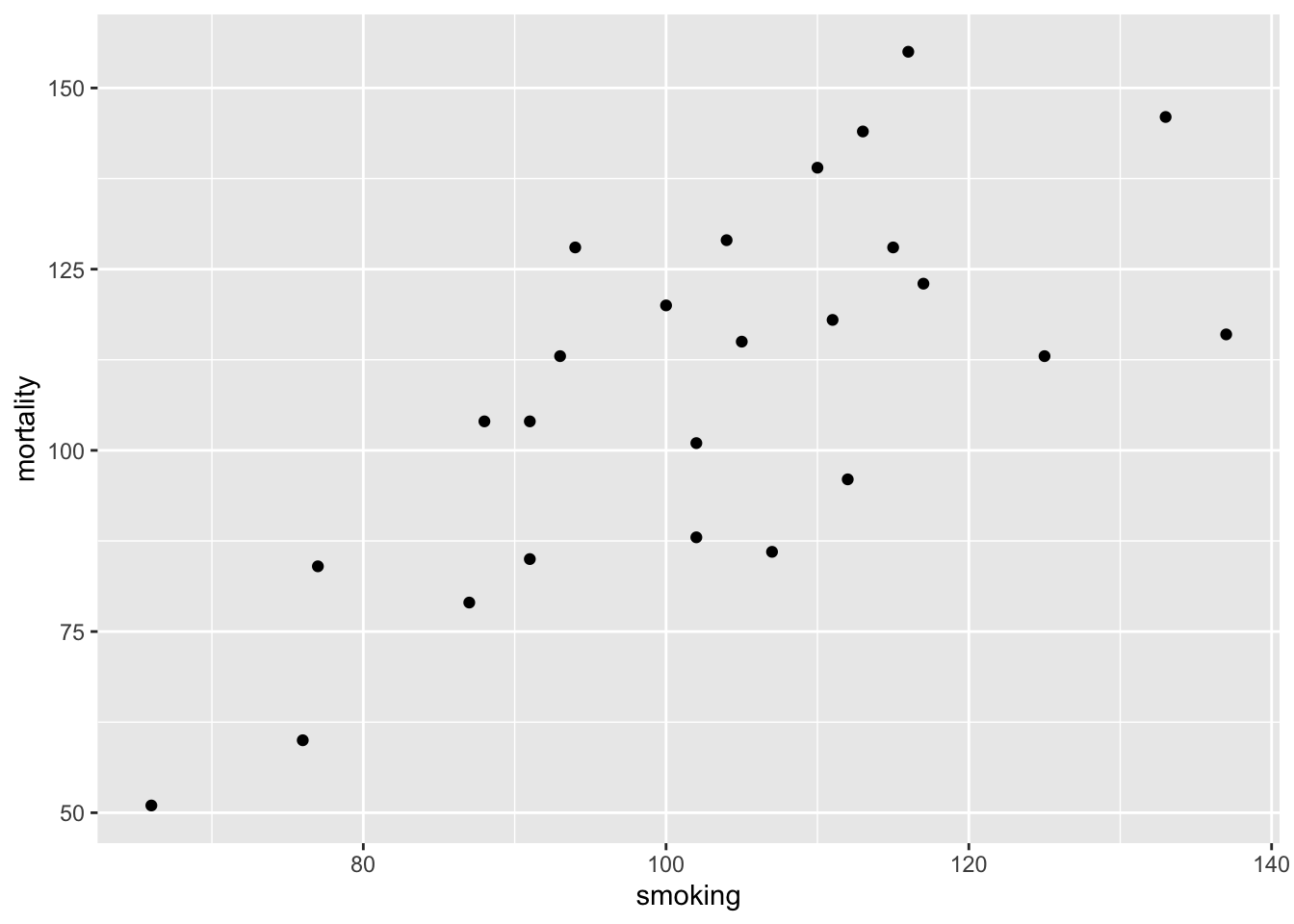
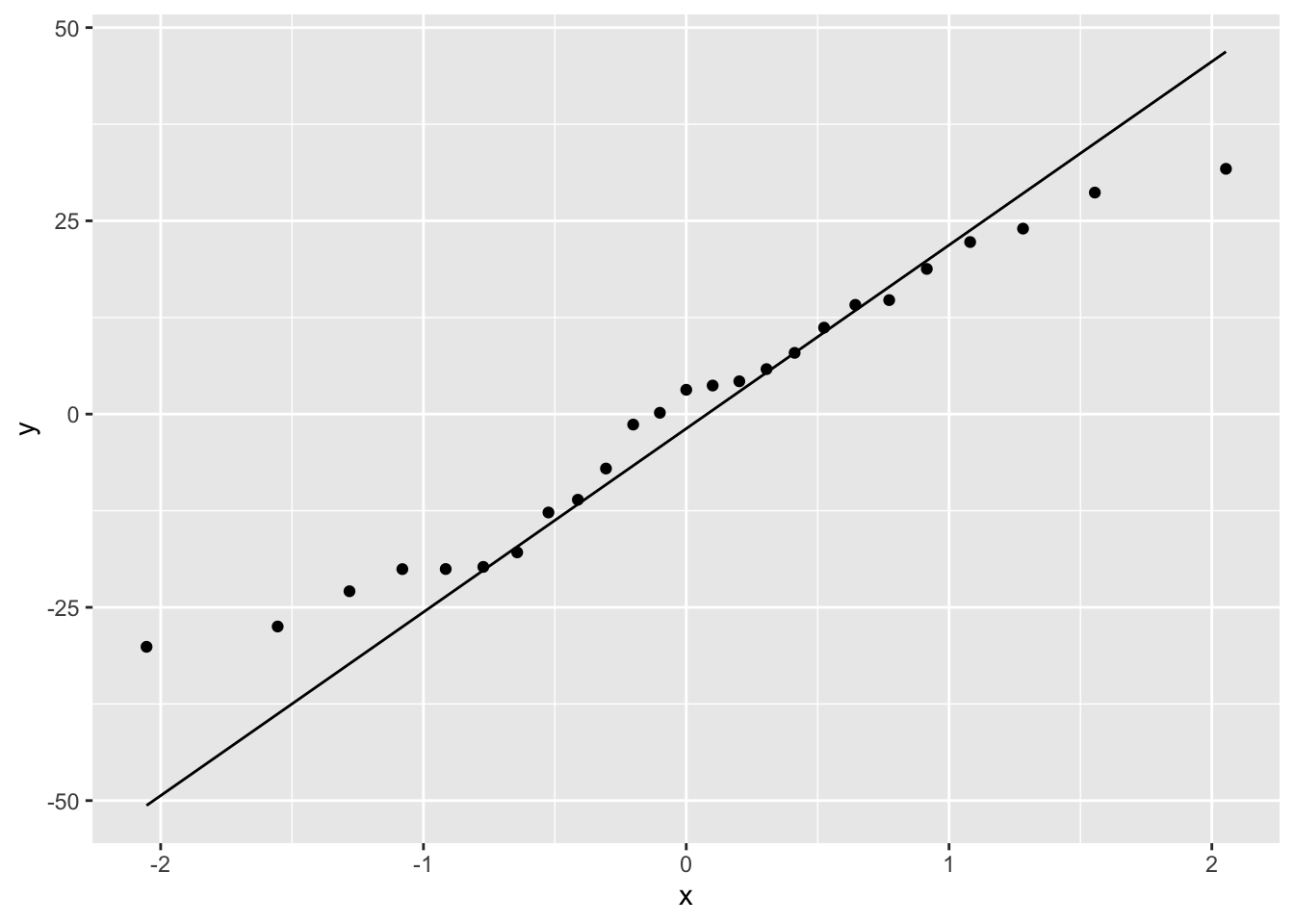
The plot does suggest that there is a positive relationship between the smoking index and mortality index.
To fit a simple linear model to these data we will use the lm() function and include our model formula mortality ~ smoking and assign the results to an object called smoke_lm .
Notice that we have not used the $ notation to specify the variables in our model formula, instead we’ve used the data = smoke argument. Although the $ notation will work (i.e. smoke$mortality ~ smoke$smoking ) it will more than likely cause you problems later on and should be avoided. In fact, we would go as far to suggest that if any function has a data = argument you should always use it. How do you know if a function has a data = argument? Just look in the associated help file.
Perhaps somewhat confusingly (at least at first) it appears that nothing much has happened, you don’t automatically get the voluminous output that you normally get with other statistical packages. In fact, what R does, is store the output of the analysis in what is known as a lm class object (which we have called smoke_lm ) from which you are able to extract exactly what you want using other functions. If you’re brave, you can examine the structure of the smoke_lm model object using the str() function.
To obtain a summary of our analysis we can use the summary() function on our smoke_lm model object.
This shows you everything you need to know about the parameter estimates (intercept and slope), their standard errors and associated t statistics and p values. The estimate for the Intercept suggests that when the relative smoking index is 0 the relative mortality rate is -2.885 ! The p value associated with the intercept tests the null hypothesis that the intercept is equal to zero. As the p value is large we fail to reject this null hypothesis. The smoking parameter estimate ( 1.0875 ) is the estimate of the slope and suggests that for every unit increase in the average number of cigarettes smoked each day the mortality risk index increases by 1.0875. The p value associated with the smoking parameter tests whether the slope of this relationship is equal to zero (i.e. no relationship). As our p value is small we reject this null hypothesis and therefore the slope is different from zero and therefore there is a significant relationship. The summary table also includes other important information such as the coefficient of determination ( R 2 ), adjusted R 2 , F statistic, associated degrees of freedom and p value. This information is a condensed form of an ANOVA table which you can see by using the anova() function.
Now let’s fit another linear model, but this time we will use the risk.group variable as our explanatory variable. Remember the risk.group variable is a factor and so our linear model will be equivalent to an ANOVA type analysis. We will be testing the null hypothesis that there is no difference in the mean mortality rate between the low , medium and high groups. We fit the model in exactly the same way as before.
Again, we can produce an ANOVA table using the anova() function
The results presented in the ANOVA table suggest that we can reject the null hypothesis (very small p value) and therefore the mean mortality rate index is different between low , medium and high risk groups.
As we did with our first linear model we can also produce a summary of the estimated parameters using the summary() function.
In the summary table the Intercept is set to the first level of risk.group ( high ) as this occurs first alphabetically. Therefore, the estimated mean mortality index for high risk individuals is 135 . The estimates for risk.grouplow and risk.groupmedium are mean differences from the intercept ( high group). So the mortality index for the low group is 135 - 57.83 = 77.17 and for the medium group is 135 - 27.55 = 107.45 . The t values and p values in the summary table are associated with testing specific hypotheses. The p value associated with the intercept tests the null hypothesis that the mean mortality index for the high group is equal to zero. To be honest this is not a particularly meaningful hypothesis to test but we can reject it anyway as we have a very small p value. The p value for the risk.grouplow parameter tests the null hypothesis that the mean difference between high and low risk groups is equal to zero (i.e. there is no difference). Again we reject this null hypothesis and conclude that the means are different between these two groups. Similarly, the p value for risk.groupmedium tests the null hypothesis that the mean difference between high and medium groups is equal to zero which we also reject.
Don’t worry too much if you find the output from the summary() function a little confusing. Its takes a bit of practice and experience to be able to make sense of all the numbers. Remember though, the more complicated your model is, the more complicated your interpretion will be. And always remember, a model that you can’t interpret is not worth fitting (most of the time!).
Another approach to interpreting your model output is to plot a graph of your data and then add the fitted model to this plot. Let’s go back to the first linear model we fitted ( smoke_lm ). We can add the fitted line to our previous plot using the ggplot2 package and the geom_smooth geom. We can easily include the standard errors by specifying the se = TRUE argument.
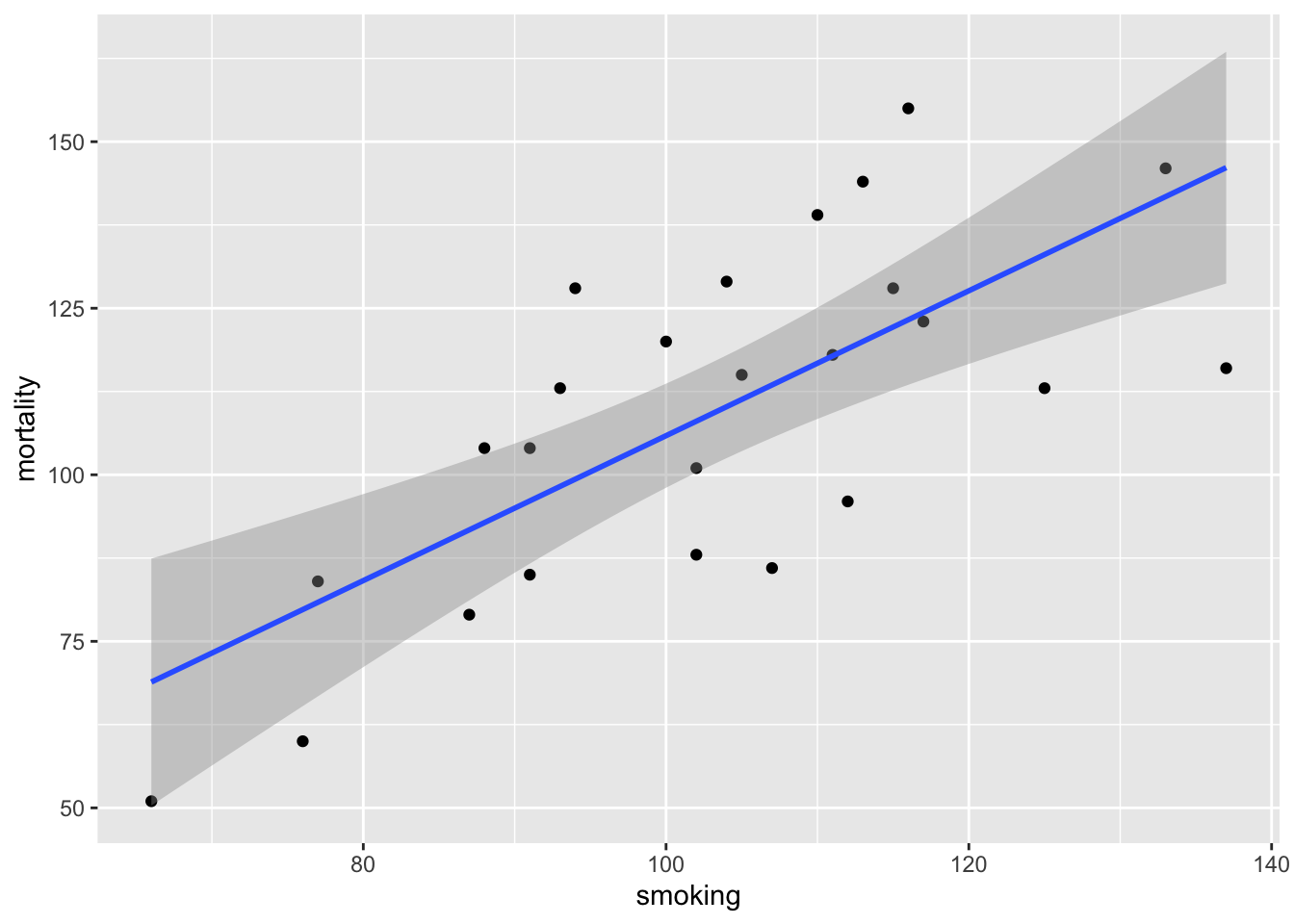
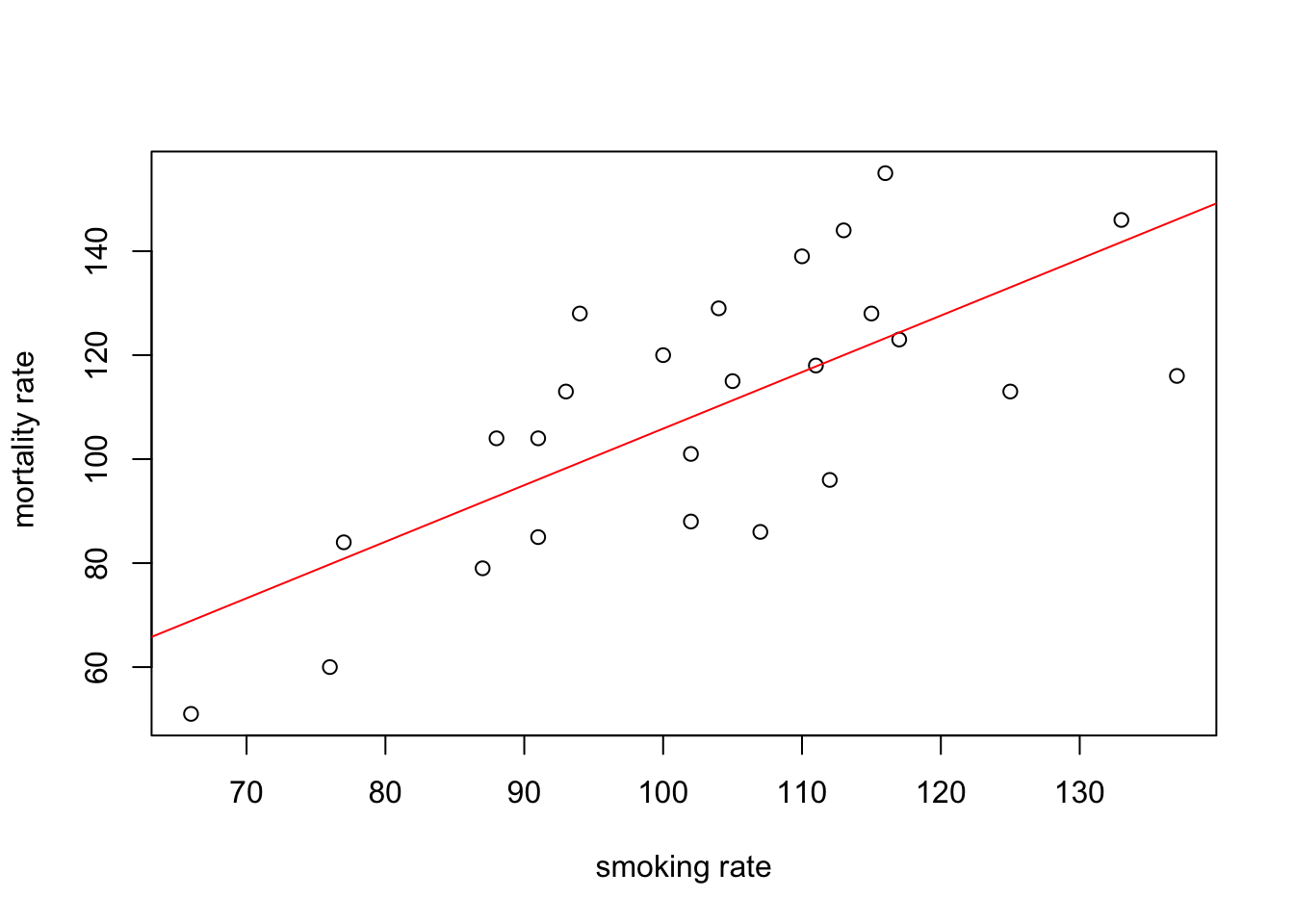
You can also do this with R’s base graphics. Note though that the fitted line extends beyond the data which is not great practice. If you want to prevent this you can generate predicted values from the model using the predict() function within the range of your data and then add these values to the plot using the lines() function (not shown).
Before we sit back and relax and admire our model (or go write that high impact paper your supervisor/boss has been harassing you about) our work is not finished. It’s vitally important to check the underlying assumptions of your linear model. Two of the most important assumption are equal variances (homogeneity of variance) and normality of residuals. To check for equal variances we can construct a graph of residuals versus fitted values. We can do this by first extracting the residuals and fitted values from our model object using the resid() and fitted() functions.
And then plot them using ggplot or base R graphics.
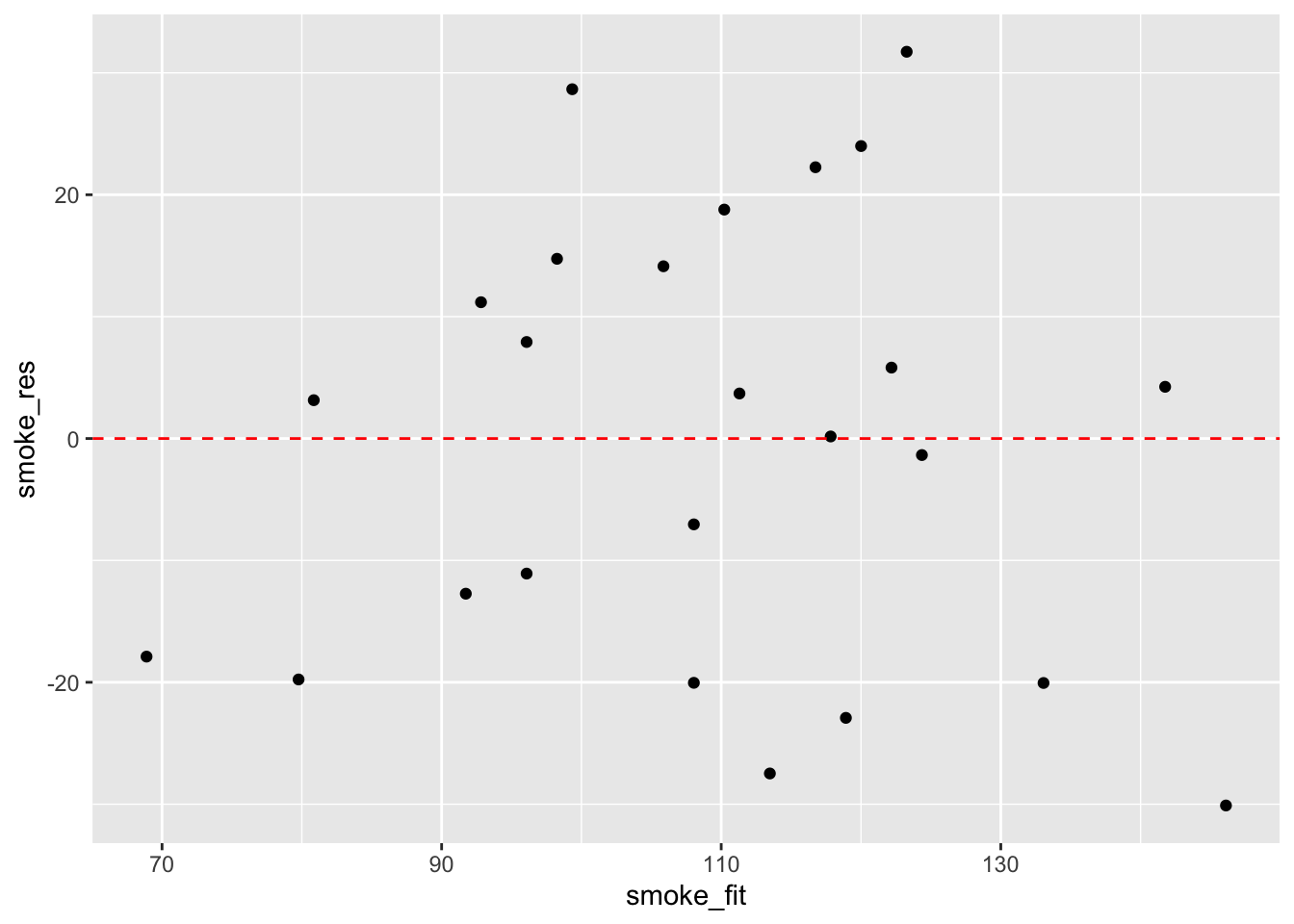
It takes a little practice to interpret these types of graph, but what you are looking for is no pattern or structure in your residuals. What you definitely don’t want to see is the scatter increasing around the zero line (red dashed line) as the fitted values get bigger (this has been described as looking like a trumpet, a wedge of cheese or even a slice of pizza) which would indicate unequal variances (heteroscedacity).
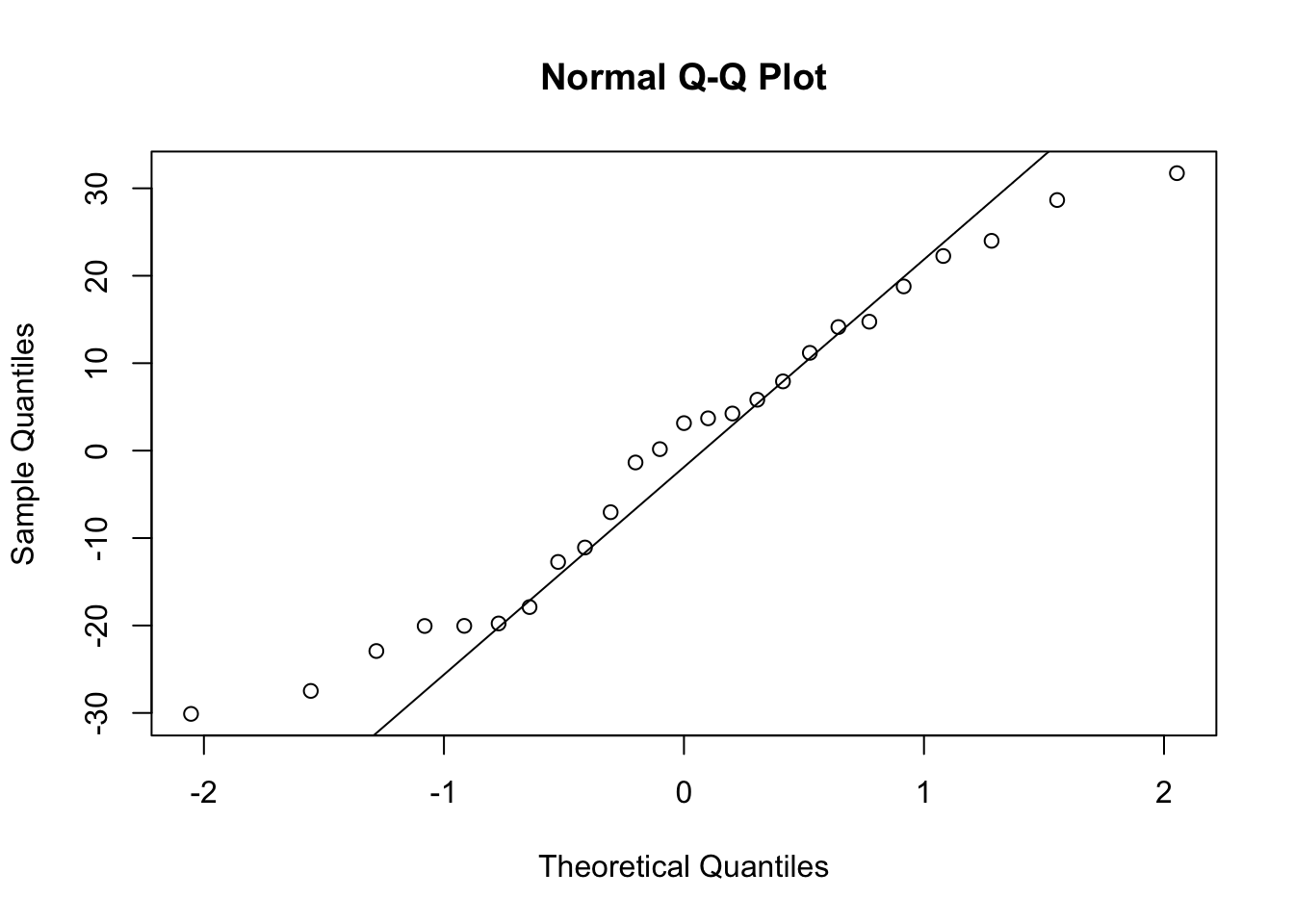
To check for normality of residuals we can use our old friend the Q-Q plot using the residuals stored in the smoke_res object we created earlier.
Or the same plot with base graphics.
Alternatively, you can get R to do most of the hard work by using the plot() function on the model object smoke_lm . Before we do this we should tell R that we want to plot four graphs in the same plotting window in RStudio using the par(mfrow = c(2,2)) . This command splits the plotting window into 2 rows and 2 columns.
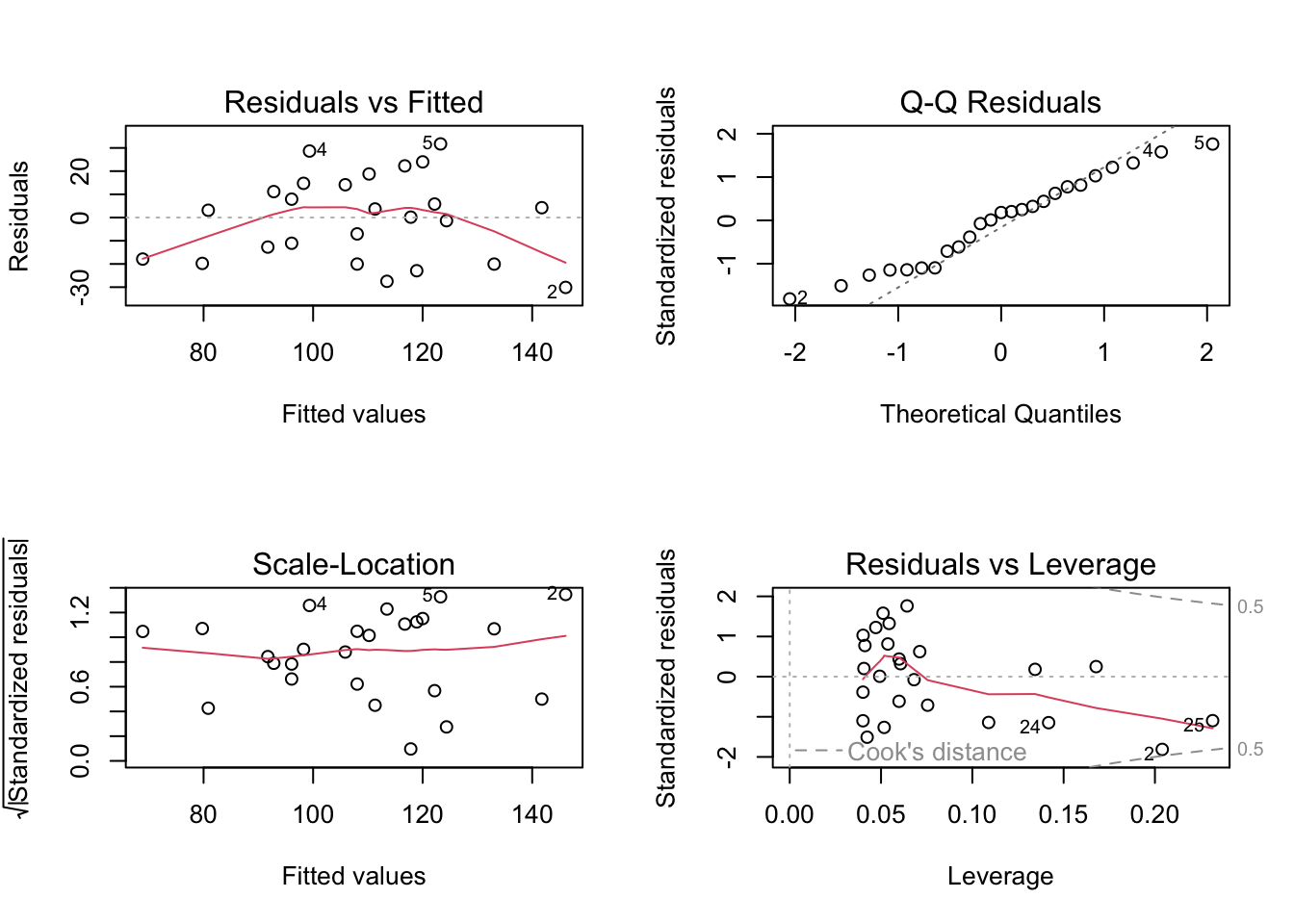
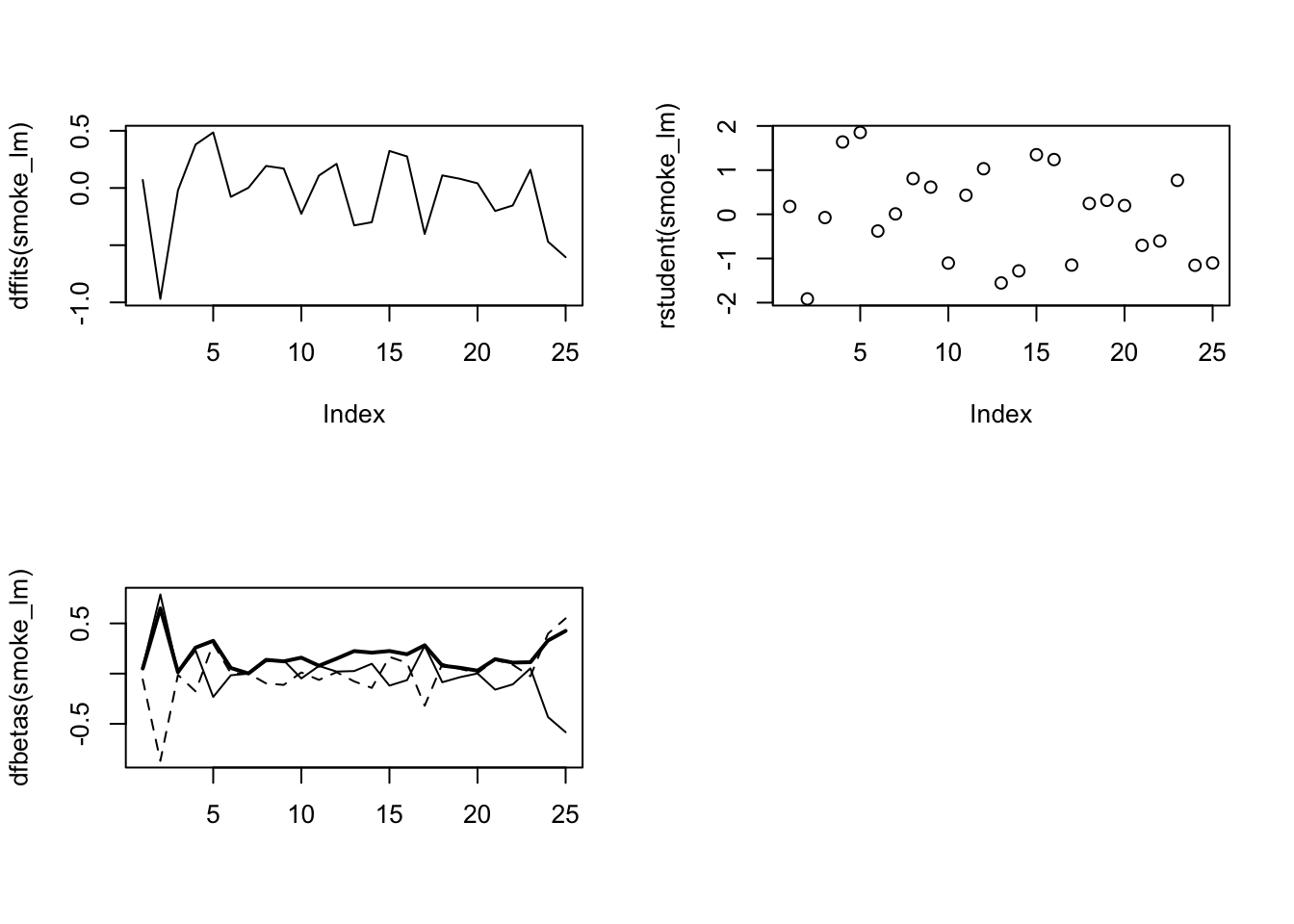
The first two graphs (top left and top right) are the same residual versus fitted and Q-Q plots we produced before. The third graph (bottom left) is the same as the first but plotted on a different scale (the absolute value of the square root of the standardised residuals) and again you are looking for no pattern or structure in the data points. The fourth graph (bottom right) gives you an indication whether any of your observations are having a large influence (Cook’s distance) on your regression coefficient estimates. Levearge identifies observations which have unusually large values in their explanatory variables.
You can also produce these diagnostic plots using ggplot by installing the package ggfortify and using the autoplot() function.
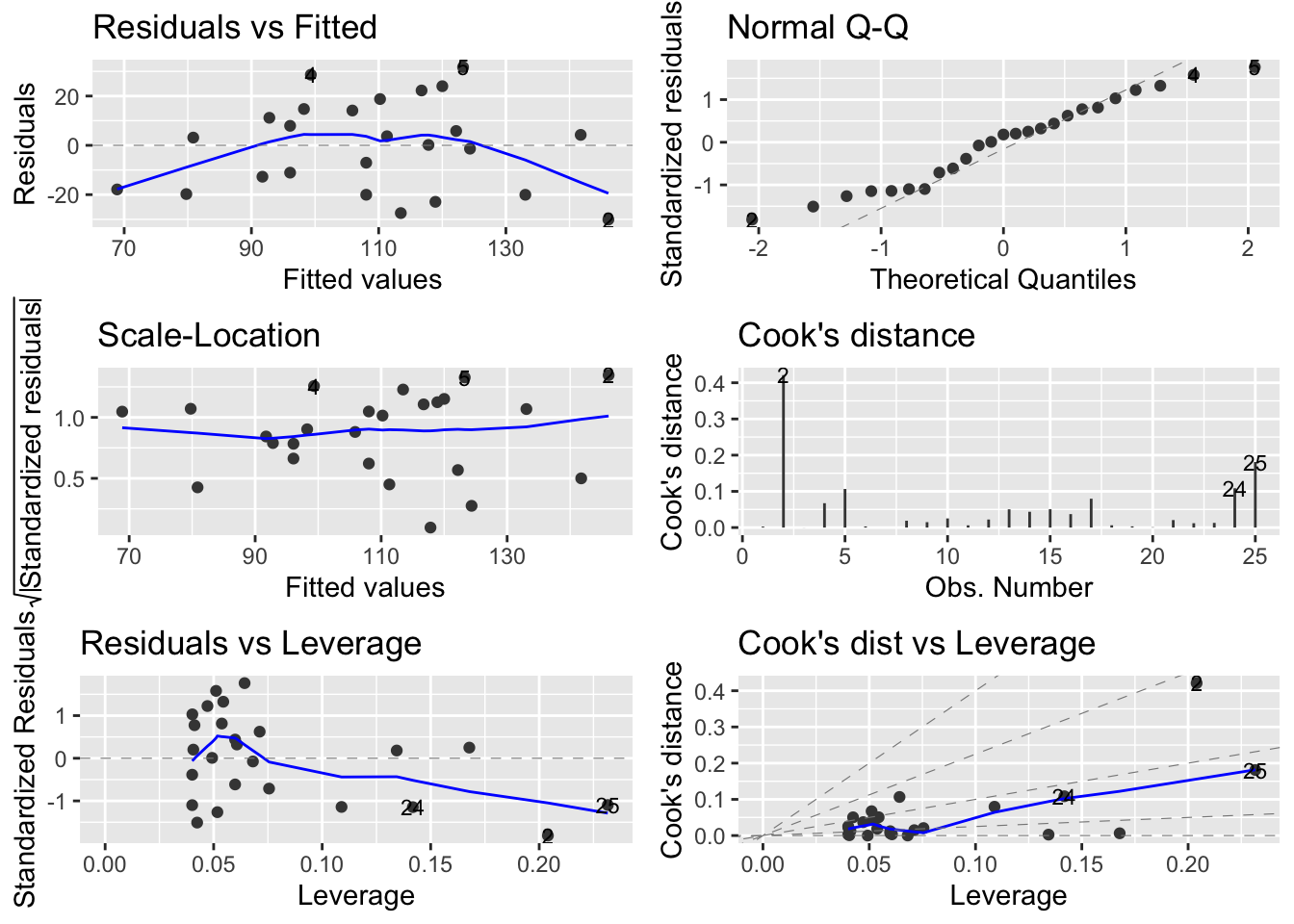
What you do about influential data points or data points with high leverage is up to you. If you would like to examine the effect of removing one of these points on the parameter estimates you can use the update() function. Let’s remove data point 2 (miners, mortality = 116 and smoking = 137) and store the results in a new object called smoke_lm2 . Note, we do this to demonstrate the use of the update() function. You should think long and hard about removing any data point(s) and if you do you should always report this and justify your reasoning.
There are numerous other functions which are useful for producing diagnostic plots. For example, rstandard() and rstudent() returns the standardised and studentised residuals. The function dffits() expresses how much an observation influences the associated fitted value and the function dfbetas() gives the change in the estimated parameters if an observation is excluded, relative to its standard error (intercept is the solid line and slope is the dashed line in the example below). The solid bold line in the same graph represents the Cook’s distance. Examples of how to use these functions are given below.

Secure Your Spot in Our PCA Online Course Starting on April 02 (Click for More Info)
Extract Residuals & Sigma from Linear Regression Model in R (3 Examples)
This tutorial shows how to return the residuals of a linear regression and descriptive statistics of the residuals in R .
Table of contents:
With that, let’s do this!
Introduction of Example Data
As a first step, I need to define some example data:
Have a look at the previous output of the RStudio console. It shows that our example data has six columns. The variable y is the outcome variable of our model and the variables x1-x5 are the predictors.
Let’s apply the summary and lm functions to estimate our linear regression model in R:
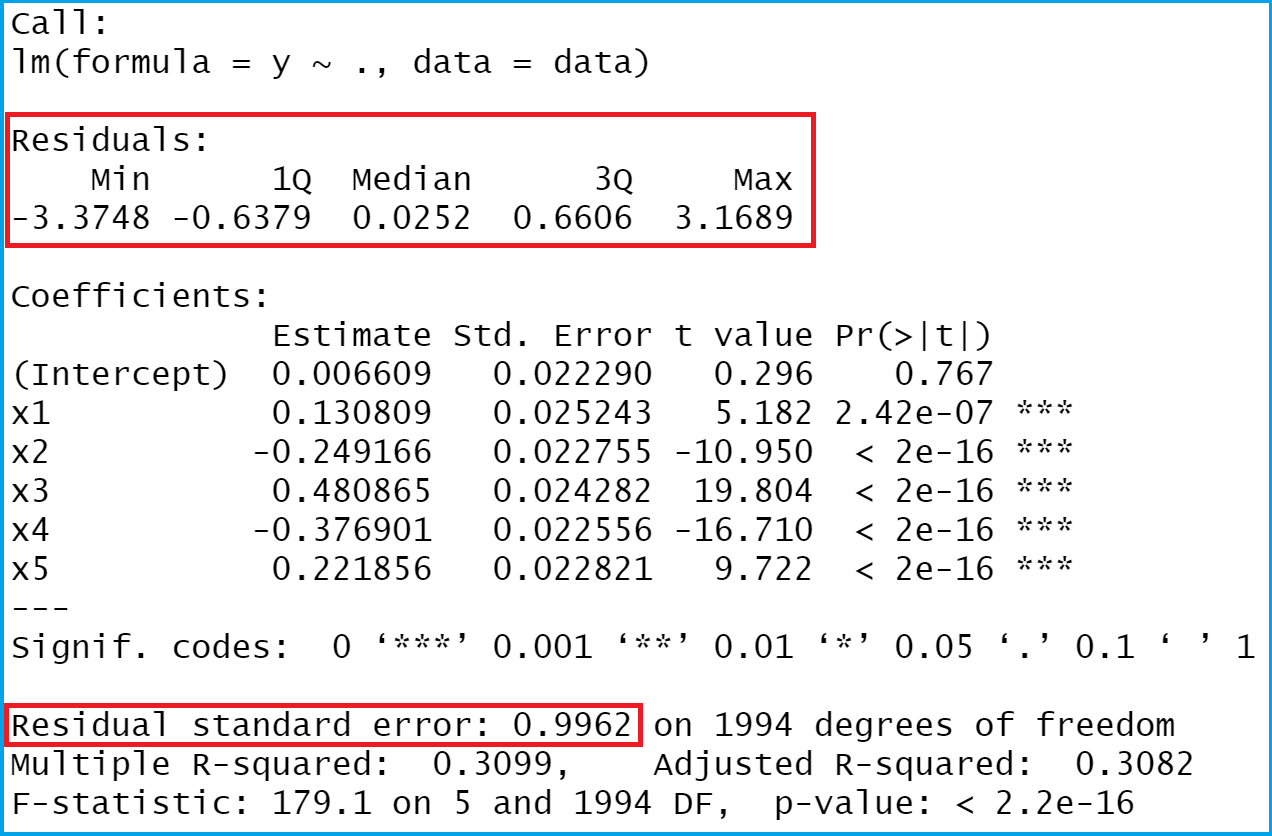
The previous Figure shows the output of our linear model. The red boxes show the values that we want to extract, i.e. the residuals and some descriptive statistics of the residuals.
Let’s do this in R!
Example 1: Extracting Residuals from Linear Regression Model
The syntax below explains how to pull out the residuals from our linear regression model.
As you can see based on the previous RStudio console output, we printed a named vector of residuals – one residual for each of the 2000 observations of our data set.
Example 2: Compute Summary Statistics of Residuals Using summary() Function
We can compute descriptive statistics of our residuals by applying the summary function to our residuals vector that we have extracted in Example 1:
The previous output is showing minimum, 1st. quartile, median, mean, 3rd quartile and the maximum value of our residuals.
Example 3: Extracting Residual Standard Error from Linear Regression Model
The R syntax below explains how to pull out the standard error of our residuals. In the terminology of the lm function, the residual standard error is called sigma:
The residual standard error of our linear model is 0.9961942.
Video & Further Resources
In case you need further info on the examples of this post, you may want to watch the following video of my YouTube channel. I’m explaining the R programming codes of this article in the video.
The YouTube video will be added soon.
Furthermore, you might want to have a look at some of the related tutorials of my homepage. I have published several articles about topics such as regression models, coding errors, and extracting data:
- Standard Error in R
- Extract Regression Coefficients of Linear Model
- All R Programming Examples
To summarize: In this article, I explained how to extract residuals from a linear model in R programming. Let me know in the comments section below, if you have additional questions.
Subscribe to the Statistics Globe Newsletter
Get regular updates on the latest tutorials, offers & news at Statistics Globe. I hate spam & you may opt out anytime: Privacy Policy .
Leave a Reply Cancel reply
Your email address will not be published. Required fields are marked *
Post Comment

I’m Joachim Schork. On this website, I provide statistics tutorials as well as code in Python and R programming.
Statistics Globe Newsletter
Get regular updates on the latest tutorials, offers & news at Statistics Globe. I hate spam & you may opt out anytime: Privacy Policy .
Related Tutorials

Variance in R (3 Examples) | Apply var Function with R Studio

Weighted Frequency Table in R (Example)

Statistics Made Easy
How to Plot lm() Results in R
You can use the following methods to plot the results of the lm() function in R:
Method 1: Plot lm() Results in Base R
Method 2: Plot lm() Results in ggplot2
The following examples shows how to use each method in practice with the built-in mtcars dataset in R.
Example 1: Plot lm() Results in Base R
The following code shows how to plot the results of the lm() function in base R:

The points in the plot represent the raw data values and the straight diagonal line represents the fitted regression line.
Example 2: Plot lm() Results in ggplot2
The following code shows how to plot the results of the lm() function using the ggplot2 data visualization package:

The blue line represents the fitted regression line and the grey bands represent the 95% confidence interval limits.
To remove the confidence interval limits, simply use se=FALSE in the stat_smooth() argument:

You can also add the fitted regression equation inside the chart by using the stat_regline_equation() function from the ggpubr package:

Additional Resources
The following tutorials explain how to perform other common tasks in R:
How to Perform Simple Linear Regression in R How to Interpret Regression Output in R The Difference Between glm and lm in R
Published by Zach
Leave a reply cancel reply.
Your email address will not be published. Required fields are marked *

IMAGES
VIDEO
COMMENTS
In this post we describe how to interpret the summary of a linear regression model in R given by summary (lm). We discuss interpretation of the residual quantiles and summary statistics, the standard errors and t statistics , along with the p-values of the latter, the residual standard error, and the F-test. Let's first load the Boston ...
Example: Interpreting Regression Output in R. The following code shows how to fit a multiple linear regression model with the built-in mtcars dataset using hp, drat, and wt as predictor variables and mpg as the response variable: #fit regression model using hp, drat, and wt as predictors. model <- lm(mpg ~ hp + drat + wt, data = mtcars)
Home » Tutorials - SAS / R / Python / By Hand Examples » Explaining the lm() Summary in R. Explaining the lm() Summary in R. Summary: Residual Standard Error: Essentially standard deviation of residuals / errors of your regression model. ... R's lm() function is fast, easy, and succinct. However, when you're getting started, that ...
The function summary.lm computes and returns a list of summary statistics of the fitted linear model given in object, using the components (list elements) "call" and "terms" from its argument, plus. the weighted residuals, the usual residuals rescaled by the square root of the weights specified in the call to lm.
In real life, most cases will not follow a perfectly straight line, so residuals are expected. In the R summary of the lm function, you can see descriptive statistics about the residuals of the model, following the same example, the red square shows how the residuals are approximately zero. How to Test if your Linear Model has a Good Fit
The model above is achieved by using the lm() function in R and the output is called using the summary() function on the model. Below we define and briefly explain each component of the model output: Formula Call. As you can see, the first item shown in the output is the formula R used to fit the data.
$\begingroup$ @godzilla For t-values, the most simple explanation is that you can use 2 (as a rule of thumb) as the threshold to decide whether or not a variable is statistically significant. Above two and the variable is statistically significant and below zero is not statistically significant. For an easy treatment of this material see Chapter 5 of Gujarati's Basic Econometrics.
In other words, we can say that a portfolio has significant exposure to a factor. R's lm() summary calculates the p-value Pr(>|t|). The smaller the p-value is, the more significant the factor is. P-value = 0.05 is a reasonable threshold. Share.
lm is used to fit linear models. It can be used to carry out regression, single stratum analysis of variance and analysis of covariance (although aov may provide a more convenient interface for these).
To understand what the coefficients are, we need to go back to what we are actually trying to do when we build a linear model. We are looking to build a generalized model in the form of y=mx+b, where b is the intercept and m is the slope of the line. Because we often don't have enough information or data to know the exact equation that exists in the wild, we have to build this equation by ...
How to interpret the output of the summary method for an lm object in R? [duplicate] (2 answers) What is the meaning of p values and t values in statistical tests? (15 answers) Closed 3 years ago. Consider a dataframe ("df") with three variables (Happiness, Smoke, Depression), where (1) Happiness (DV) = continuous measure of happiness on 1-10 ...
Linear model summaries as HTML table. The sjt.lm function prints summaries of linear models (fitted with the lm function) as nicely formatted html-tables. Before starting, sample data is loaded and sample models are fitted: # sample data data (efc) # set variable labels efc <- set_var_labels (efc, get_var_labels (efc)) # fit first model fit1 ...
The lm () function in R is used to fit linear regression models. This function uses the following basic syntax: lm (formula, data, …) where: formula: The formula for the linear model (e.g. y ~ x1 + x2) data: The name of the data frame that contains the data. The following example shows how to use this function in R to do the following:
Step 1: Load the data into R. Follow these four steps for each dataset: In RStudio, go to File > Import dataset > From Text (base).; Choose the data file you have downloaded (income.data or heart.data), and an Import Dataset window pops up.In the Data Frame window, you should see an X (index) column and columns listing the data for each of the variables (income and happiness or biking, smoking ...
The function summary.lm computes and returns a list of summary statistics of the fitted linear model given in object, using the components (list elements) "call" and "terms" from its argument, plus. residuals. the weighted residuals, the usual residuals rescaled by the square root of the weights specified in the call to lm.
The main function for fitting linear models in R is the lm() function (short for linear model!). The lm() function has many arguments but the most important is the first argument which specifies the model you want to fit using a model formula which typically takes the general form: response variable ~ explanatory variable(s)
This tutorial explains how to extract regression coefficients from the lm() function in R, including several examples.
Details on formatting of the coefficients in the summary: It is not obvious where exactly the formatting happens: The summary() method for lm objects returns an object of class summary.lm which has its own print() method which in turn calls printCoefmat(). The latter is the function that does the actual formatting.
Algebraically, the equation for a simple regression model is: y^i = β^0 + β^1xi +ε^i where ε ∼ N(0, σ^2) y ^ i = β ^ 0 + β ^ 1 x i + ε ^ i where ε ∼ N ( 0, σ ^ 2) We just need to map the summary.lm() output to these terms. To wit:
This tutorial shows how to return the residuals of a linear regression and descriptive statistics of the residuals in R. Table of contents: 1) Introduction of Example Data. 2) Example 1: Extracting Residuals from Linear Regression Model. 3) Example 2: Compute Summary Statistics of Residuals Using summary () Function.
Example 4: Using summary () with Regression Model. The following code shows how to use the summary () function to summarize the results of a linear regression model: #define data. df <- data.frame(y=c(99, 90, 86, 88, 95, 99, 91), x=c(33, 28, 31, 39, 34, 35, 36)) #fit linear regression model.
You may check broom to convert "the messy output of built-in functions in R, such as lm [..] and turns them into tidy data frames". - Henrik May 21, 2015 at 11:06
How to Plot lm () Results in R. You can use the following methods to plot the results of the lm () function in R: Method 1: Plot lm () Results in Base R. #add fitted regression line to scatterplot. Method 2: Plot lm () Results in ggplot2. geom_point() +. stat_smooth(method = "lm")